Mini review 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Important Structural Insights of the 2019-nCoV Spike Protein
*Corresponding author:Mir Monir Hossain, Department of Chemistry, Cleveland State University (CSU), Cleveland, Ohio 44115, USA and Department of Pharmacy, University of Science & Technology Chittagong (USTC), Foy’s Lake, Chittagong 4202, Bangladesh
Received:December 13, 2022; Published:December 16, 2022
DOI: 10.34297/AJBSR.2022.17.002386
Abstract
Rapid global outbreak of novel coronavirus (2019-nCoV) showed a fearful pandemic state for which a public health emergency had declared worldwide. This lethal virus exploits the glycosylated spike (S) trimers to get entry into the host cells and thereby shows its lethality in human to human. Wonderful conformational flexibility of S proteins makes easy exposure of its specific receptorbinding site and promptly undergoes complete structural rearrangement to drive fusion of viral and cellular membranes. Thus, the covid spike (S) glycoprotein is a main target of interest for the fruitful development of therapeutic antibodies, vaccines, as well as diagnostics against 2019-nCoV.
Keywords: 2019-nCoV, Spike proteins, Cryo-EM, Conformation
Introduction
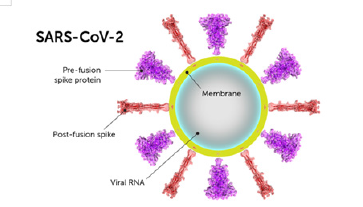
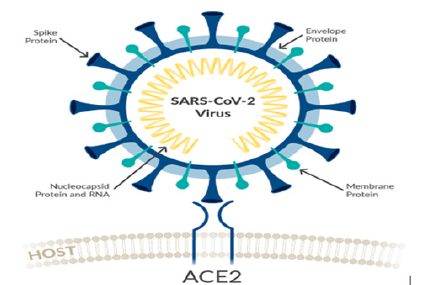
Structurally the novel coronavirus (2019-nCoV or SARS-CoV-2) surrounded by a lipid bilayer through which spike (S) protein trimers protrude [1]. Heavily glycosylated S trimers bind with angiotensin-converting enzyme 2 (ACE2) to modulate the entry of virions into host cells [2-4]. The S is class I trimeric fusion protein which can exist in a metastable conformation and shows a form of structural rearrangement to fuse viral membrane with targeted cell membrane [5,6]. The receptor-binding domain (RBD) of S protein exhibits hinge-like conformational movements to interact with a host cell ACE2 receptor that transiently remain in hide or expose state. These states are known as “down” conformation and “up” conformation, where “down” corresponds to the receptorinaccessible state and “up” denotes receptor- accessible state, which found less stable [7,8]. Based on such unique role of S trimer, it can be a best focus for antibody target, hence proper characterization of S structure will give us atomic-level insights to lead us vaccine development and design (Figure 1).
Discussion
Scientists through their intellectual and brilliant research, have already measured the cryo-electron microscopy (Cryo-EM) structure of 2019-nCoV spike (S) trimer in both pre-fusion and post-fusion conformations [9]. They also explained the prominent structural and bio-physical outcomes which support that spike protein binds to ACE2 with higher affinity than S protein of severe acute respiratory syndrome coronavirus (SARS-CoV).
Moreover, they evaluated various known SARS-CoV receptorbinding domains (RBD)-specific monoclonal antibodies and observed that they do not have specific binding to 2019-nCoV S protein, which supports the cross-reactivity of antibody may show limitations between this two RBDs. The total structural conformations study by the Cryo-EM showed that, the S of 2019- nCoV is greatly like the SARS-CoV S. But the main dissimilarities between these two S protein structures lie in the position of the RBDs of their respective down conformations. Mostly, the RBD of SARS-CoV packs tightly in the down conformation against the N-terminal domain (NTD) of nearest protomer. Whereas the RBD of 2019-nCoV in their down conformation remains angled close to the central cavity of the protein. Despite such observed conformational disparity, both proteins share a high sense of structural similarity in the case of their respective NTDs, RBDs, subdomains 1 and 2.
Latest findings reveal that both SARS-CoV S and 2019-nCoV S share the same functional host cell receptor which is ACE2. Hence the researchers used surface plasmon resonance (SPR) to show the binding kinetics of interaction of human ACE2 and 2019-nCoV S. In SPR technique they found that, this receptor binds with 2019-nCoV S in such a way which is 10 to 20 times higher than ACE2 binding with the S of SARS-CoV. This high affinity of 2019-nCoV S for human ACE2 may be a good reason why 2019-nCoV can easily spread from human to human [10]. Still vigorous studies are required to support this possibility.
In addition, by negative-stain EM it was observed that, complex from ACE2 bound to the 2019-nCoV S ectodomain, highly like the complex obtained from the interaction between SARS-CoV S and ACE2 that also observed by cryo-EM [11,12]. From such structural homology and same receptor usage, scientists also used biolayer interferometry (BLI) to test SARS-CoV RBD-directed three monoclonal antibodies (mAbs) (namely: m396, S230 and 80R) for cross-reactivity in case of 2019-nCoV RBD. But they didn’t get any binding to the SARS-CoV RBD for any of these three tested mAbs. This lacking binding leads that SARS-directed mAbs would not importantly be cross-reactive. For this reason, the future efforts of antibody isolation and therapeutic design will benefit from using 2019-nCoV S proteins as probes (Figure 2).
Conclusion
The pace of mutation of novel coronavirus is very high in compared to the discovery of effective immunogens for vaccination and antiviral drugs. This is the main cause of the global threat for another pandemic in our total healthcare system. Thus, in depth study of the atomic-level structure of 2019-nCoV spike will prompt scientists to design and develop of effective antiviral therapeutics as well as vaccines.
References
- Neuman B W, Buchmeier MJ (2016) Supramolecular architecture of the coronavirus particle. Adv Virus Res 96: 1-27.
- Walls A C, Young Jun Park, M Alejandra Tortorici, Abigail Wall, Andrew T McGuire, et al. (2020) Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181: 281-292.
- Hoffmann M, Hannah Kleine Weber, Simon Schroeder, Nadine Krüger, Tanja Herrler, et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271-280.
- Shang, J, Gang Ye, Ke Shi, Yushun Wan, Chuming Luo, et al. (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581: 221-224.
- F Li (2016) Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol 3(1): 237-261.
- Berend Jan Bosch, Ruud van der Zee, Cornelis A M de Haan, Peter J M Rottier (2003) The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77(16): 8801-8811.
- Miao Gui, Wenfei Song, Haixia Zhou, Jingwei Xu, Silian Chen, et al. (2017) Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 27(1): 119-129.
- Jesper Pallesen, Nianshuang Wang, Kizzmekia S Corbett, Daniel Wrapp, Robert N Kirchdoerfer, et al. (2017) Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci USA 114(35): E7348-E7357.
- Daniel Wrapp, Nianshuang Wang, Kizzmekia S Corbett, Jory A Goldsmith, Ching Lin Hsieh, et al. (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367(6483): 1260-1263.
- Jasper Fuk Woo Chan, Shuofeng Yuan, Kin Hang Kok, Kelvin Kai Wang To, Hin Chu, et al. (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395(10223): 514-523.
- Robert N Kirchdoerfer, Nianshuang Wang, Jesper Pallesen, Daniel Wrapp, Hannah L Turner, et al. (2018) Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep 8(1): 15701.
- Wenfei Song, Miao Gui, Xinquan Wang, Ye Xiang (2018) Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLOS Pathog 14(8): e1007236.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.